library(readr)
library(lubridate)
library(stringr)
library(tidyr)
library(dplyr)
library(ggplot2)Plotting Great Lakes water level data using R - a 2025 update
They moved the data
This post is an update of a post originally done back in 2018. The location and format for data downloading has changed.
The past few years we have seen record or near record high water levels in all of the Great Lakes. Recently, the levels have dropped. While plots of historical water levels are available on the web, I thought it would be interesting create my own plots using R. In this post I will summarize the steps for using R to:
- download raw historical water level data for the Great Lakes using a few different methods,
- process the raw data to get it into shape for time series plots,
- create time series plots of monthly water levels using
ggplot.
Getting monthly water level data
We will need a few libraries.
You can download CSV files from NOAA containing monthly water levels for all of the Great Lakes at the following location:
This URL has buttons for downloading long term monthly mean water levels in units desired. The links are:
- https://lre-wm.usace.army.mil/ForecastData/WaterLevelData/GLHYD_data_english.csv
- https://lre-wm.usace.army.mil/ForecastData/WaterLevelData/GLHYD_data_metric.csv
I’ve already downloaded it to a local location to avoid repeated downloads while writing this post.
#data_raw_loc <- "https://lre-wm.usace.army.mil/ForecastData/WaterLevelData/GLHYD_data_metric.csv"
data_raw_loc <- "data_water_level/raw/GLHYD_data_metric.csv"
data_loc <- "data_water_level"Before reading it into an R dataframe, let’s look at the file format.
# Coordinated Monthly Mean Lakewide Average Water Levels,,,,,,
# Period of record: 1918-2023,,,,,,
"# Units: meters, IGLD 1985",,,,,,
"# Calculated using the coordinated gage network, consisting of:",,,,,,
"# Superior: Marquette and Point Iroquois, MI; Duluth, MN; Michipicoten and Thunder Bay, Ontario",,,,,,
"# Michigan-Huron: Harbor Beach, Mackinaw City and Ludington, MI; Milwaukee, WI; Thessalon and Tobermory, Ontario",,,,,,
"# St. Clair: St. Clair Shores, MI and Belle River, Ontario",,,,,,
"# Erie: Toledo and Cleveland, OH; Port Stanley and Port Colborne, Ontario",,,,,,
"# Ontario: Oswego and Rochester, NY; Cobourg, Port Weller, Toronto, and Kingston, Ontario",,,,,,
#,,,,,,
# Last modified March 2024 Contact: Deanna.C.Fielder@usace.army.mil,,,,,,
#,,,,,,
month,year,Superior,Michigan-Huron,St. Clair,Erie,Ontario
jan,1918,183.25,176.71,174.59,173.9,74.74
feb,1918,183.2,176.73,174.74,173.82,74.72
mar,1918,183.17,176.8,174.74,174.01,74.92
apr,1918,183.14,176.89,174.84,174.02,75.1
may,1918,183.22,176.99,175,173.98,75.09
jun,1918,183.34,177.07,175.14,174.1,75.06
jul,1918,183.4,177.07,175.17,174.12,74.99A few things to note:
- The top twelve lines are metadata,
- Data goes back to 1918,
- Data is in wide format in that each lake is in its own column.
I used the readr package to read in this csv file. Notice that there is a single column for Lakes Michigan and Huron as they are actually just one big lake that we divide at the Straits of Mackinac - see https://www.glerl.noaa.gov/res/straits/.
mean_lake_level_raw <- read_csv(data_raw_loc, skip = 12)Data reshaping
In order to facilitate plotting with ggplot2, we are going to need to reshape this data into long format. Each row will be a single monthly reading and there will be a month column and a lake column. Let’s use the tidyr::pivot_longer function.
mean_lake_level_long <- pivot_longer(mean_lake_level_raw, names_to = "lake", values_to = "level_m", cols = 3:7)
rm(mean_lake_level_raw)Take a peek at the data in long format.
head(mean_lake_level_long)# A tibble: 6 × 4
month year lake level_m
<chr> <dbl> <chr> <dbl>
1 jan 1918 Superior 183.
2 jan 1918 Michigan-Huron 177.
3 jan 1918 St. Clair 175.
4 jan 1918 Erie 174.
5 jan 1918 Ontario 74.7
6 feb 1918 Superior 183. The months are three character strings. Let’s create a date column that we can use for joining the individual tables as well as proper sorting.
mean_lake_level_long$date <- as.POSIXct(str_c(mean_lake_level_long$month, " 1, ",
mean_lake_level_long$year),
format="%b %d, %Y")
mean_lake_level_long <- mean_lake_level_long |>
arrange(lake, date)Check out our work.
head(mean_lake_level_long)# A tibble: 6 × 5
month year lake level_m date
<chr> <dbl> <chr> <dbl> <dttm>
1 jan 1918 Erie 174. 1918-01-01 00:00:00
2 feb 1918 Erie 174. 1918-02-01 00:00:00
3 mar 1918 Erie 174. 1918-03-01 00:00:00
4 apr 1918 Erie 174. 1918-04-01 00:00:00
5 may 1918 Erie 174. 1918-05-01 00:00:00
6 jun 1918 Erie 174. 1918-06-01 00:00:00Alternative data source from GLCC
The Great Lakes Coordination Committee (US and CA) does what it sounds like it does.
The Coordinating Committee on Great Lakes Basic Hydraulic and Hydrologic Data (Coordinating Committee) is a collaboration of the Governments of the United States and Canada for the purpose of agreeing upon the basic hydraulic, hydrologic and vertical control data that is required to manage the Great Lakes and St. Lawrence River.
They also maintain a repository of Great Lakes water level data at https://www.greatlakescc.org/en/coordinating-committee-products-and-datasets/. There are CSV files stored on Google Drive for each lake containing mean monthly water levels since 1918. These are in a different wide format than the data above. Let’s look at the data for Lake Michigan-Huron and compare to the data we just downloaded. Here is what the raw data looks like:
# NAME: LAKE MICHIGAN-HURON MONTHLY MEAN LEVELS
# UNITS: metres (m) (DATUM: International Great Lakes Datum of 1985)
# PERIOD OF RECORD: 1918 TO 2024
# DESCRIPTION: Water levels are measured by the National Oceanic and Atmospheric Administration and the
# Canadian Department of Fisheries and Oceans. Mean levels are computed as an average
# of a network of gauges and coordinated under the auspices of the Coordinating Committee on
# Great Lakes Basic Hydraulic and Hydrologic Data www.greatlakescc.org
# # All data for the previous 12 months should be considered provisional.
# note: -9999 means that no data is available for that date
Year, Jan, Feb, Mar, Apr, May, Jun, Jul, Aug, Sep, Oct, Nov, Dec
1918,176.71,176.73,176.8,176.89,176.99,177.07,177.07,177.01,176.94,176.83,176.82,176.78
1919,176.74,176.68,176.68,176.77,176.89,176.92,176.88,176.83,176.72,176.66,176.62,176.55
1920,176.5,176.47,176.49,176.63,176.68,176.73,176.79,176.76,176.73,176.65,176.57,176.5
1921,176.44,176.42,176.42,176.54,176.63,176.64,176.6,176.54,176.5,176.44,176.35,176.34
1922,176.27,176.24,176.28,176.43,176.57,176.63,176.67,176.63,176.56,176.46,176.35,176.25 A few things to note:
- The top nine lines are metadata,
- Data goes back to 1918,
- Data is in wide format in that each month (instead of lake) is in its own column.
We need to combine the separate lake files and do some reshaping and column manipulation. Again, I’ve already downloaded the five data files to a local folder.
data_cc_raw_loc <-"data_water_level/raw/"
gl_data_raw = list()
gl_data_long = list()
# regex pattern for filename matching
file_pattern <- "\\w+_MonthlyMeanWaterLevels_1918to2024\\.csv$"
gl_files <- list.files(path = data_cc_raw_loc, pattern = file_pattern)
# Loop over the list of matched filenames
for (i in seq_along(gl_files)) {
filename <- paste0(data_cc_raw_loc, gl_files[i])
gl_data_raw[[i]] <- read_csv(file = filename, skip = 9)
# Which lake?
lake <- str_match(filename, "(Lake\\w+)_Month")[2]
# Pivot to long format
gl_data_long[[i]] <- pivot_longer(gl_data_raw[[i]], names_to = "month", values_to = "level_m", cols = 2:13)
# Add a lake identifier column
gl_data_long[[i]]$lake <- lake
# Add date based on month and year
gl_data_long[[i]]$date <- as.POSIXct(str_c(gl_data_long[[i]]$month, " 1, ",
gl_data_long[[i]]$Year),
format="%b %d, %Y")
}
# Bind all the rows together from the list of dataframes
mean_lake_level_cc_long <- bind_rows(gl_data_long)
# Column renaming and factor seting
mean_lake_level_cc_long <- mean_lake_level_cc_long %>%
rename(year = Year)
mean_lake_level_cc_long$lake <- factor(mean_lake_level_cc_long$lake,
levels = c("LakeSuperior", "LakeMichiganHuron", "LakeStClair", "LakeErie", "LakeOntario"))This data goes through 2024 whereas the one from the NOAA site hasn’t been updated yet for 2024. Let’s check to see if data is the same through 2023.
level_m_noaa <- mean_lake_level_long %>%
select(level_m)
level_m_cc <- mean_lake_level_cc_long %>%
filter(year < 2024) %>%
select(level_m)
max_abs_diff <- max(abs(level_m_noaa$level_m - level_m_cc$level_m), na.rm = TRUE)
assertthat::assert_that(max_abs_diff == 0)[1] TRUEConfirms that the data is identical from the two sources.
Lake level plotting
If we plot all the lake level time series on one plot, we can see the differences in levels between lakes but the intra-lake variation is hidden by the scale.
ggplot(mean_lake_level_cc_long) + geom_line(aes(x=date, y = level_m, colour = lake)) +
ylab("Water level (m)")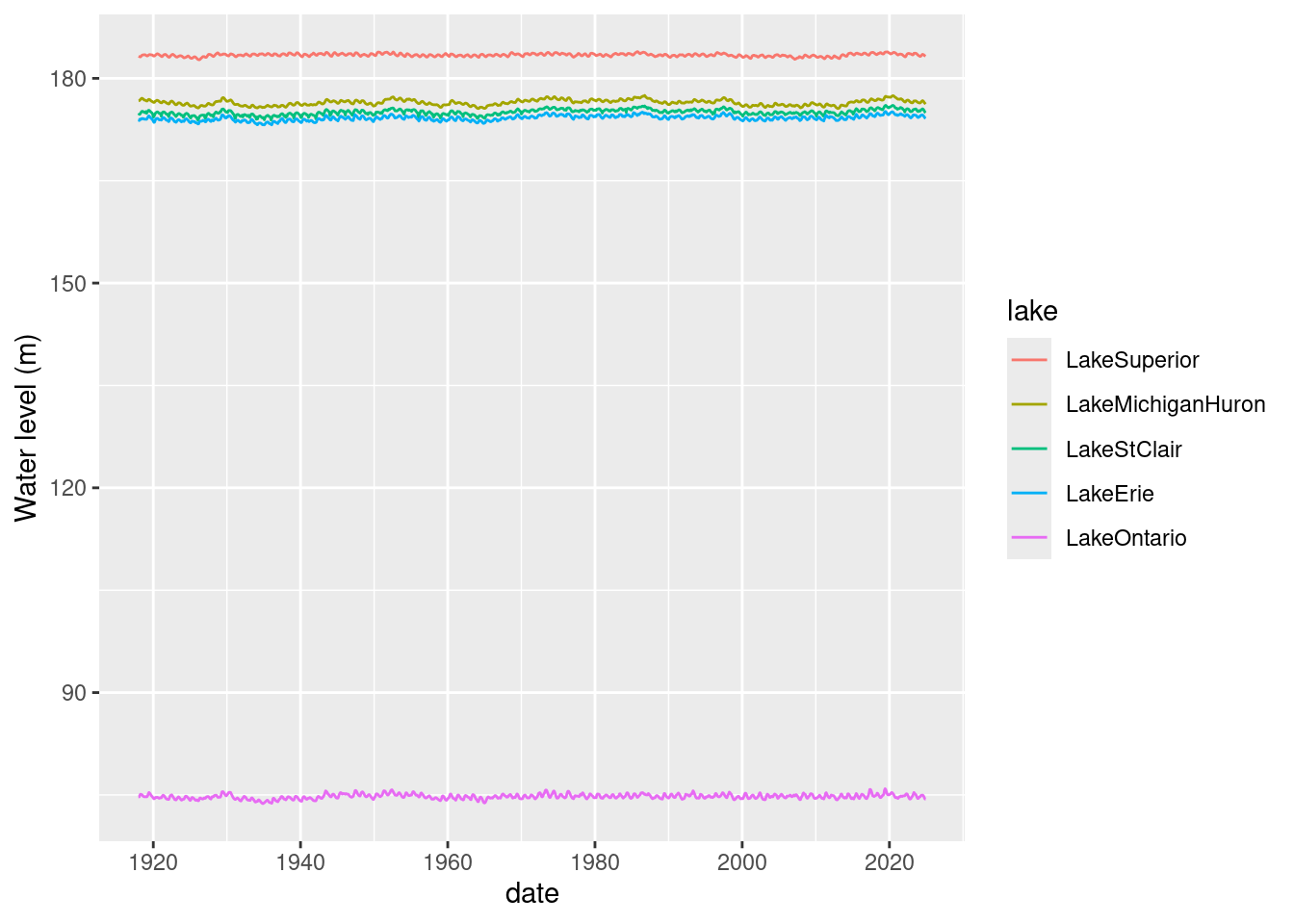
Let’s find the latest month for which we have data and save a version of this dataframe and tag it by the month. Note the use of pull() to convert the the resulting 1x1 dataframe into a value.
last_month_d <- mean_lake_level_cc_long %>%
filter(!is.na(level_m)) %>%
select(date) %>%
summarize(whichmonth = max(date)) %>%
pull()
last_month_d[1] "2024-12-01 EST"Save as an rds file.
rdsname <- file.path(data_loc, paste0("lake_level_", year(last_month_d), strftime(last_month_d,"%m"), ".rds"))
saveRDS(mean_lake_level_long, rdsname)Plotting monthly water level data
Let’s compute a bunch of overall historical statistics and include a few as reference lines on the plots. I will use dplyr for the stats.
lake_level_stats <- mean_lake_level_cc_long %>%
group_by(lake) %>%
summarize(
mean_level = mean(level_m, na.rm = TRUE),
min_level = min(level_m, na.rm = TRUE),
max_level = max(level_m, na.rm = TRUE),
p05_level = quantile(level_m, 0.05, na.rm = TRUE),
p95_level = quantile(level_m, 0.95, na.rm = TRUE),
sd_level = sd(level_m, na.rm = TRUE),
cv_level = sd_level / mean_level
)
lake_level_stats# A tibble: 5 × 8
lake mean_level min_level max_level p05_level p95_level sd_level cv_level
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 LakeSupe… 183. 183. 184. 183. 184. 0.203 0.00110
2 LakeMich… 176. 176. 178. 176. 177. 0.406 0.00230
3 LakeStCl… 175. 174. 176. 174. 176. 0.395 0.00226
4 LakeErie 174. 173. 175. 174. 175. 0.369 0.00212
5 LakeOnta… 74.8 73.7 75.9 74.2 75.3 0.343 0.00459Let’s try to combine time series with lines from the stats dataframe just created. There are a few approaches to doing this. One way is to add geom_hline objects to the plot. See the following posts for some good ideas on using geom_hline(), adding the reference lines to the legend and controlling their style and color, and controlling the order of plots in a faceted grid.
- https://stackoverflow.com/questions/11846295/how-to-add-different-lines-for-facets
- https://forum.posit.co/t/scale-linetype-manual-not-working/70225
- https://stackoverflow.com/questions/75882636/how-to-specify-geom-hline-color-and-linetype-in-ggplot-legend
- https://stackoverflow.com/questions/75450315/r-facet-grid-order-not-following-levels-with-plotmath
# Create a vector of line style assignments
LINES <- c("mean" = "solid", "5%" = "dotted", "95%" = "dotted")
# Need to explicitly specify order in facet_grid even though levels already set in dataframe
ggplot(mean_lake_level_cc_long, aes(x = date, y = level_m, colour = lake)) +
facet_grid(factor(lake, levels = c("LakeSuperior", "LakeMichiganHuron", "LakeStClair", "LakeErie", "LakeOntario")) ~ ., scales = "free") +
geom_hline(data = lake_level_stats, aes(linetype = "mean", yintercept=mean_level)) +
geom_hline(data = lake_level_stats, aes(linetype = "5%", yintercept=p05_level)) +
geom_hline(data = lake_level_stats, aes(linetype = "95%", yintercept=p95_level)) +
geom_line() +
scale_linetype_manual(values = LINES) +
ylab("Lake Level (m)") +
theme(strip.text.y = element_text(size = 16),
axis.text.x = element_text(size = 14),
axis.text.y = element_text(size = 10),
axis.title.x = element_text(size=18),
axis.title.y = element_text(size=18))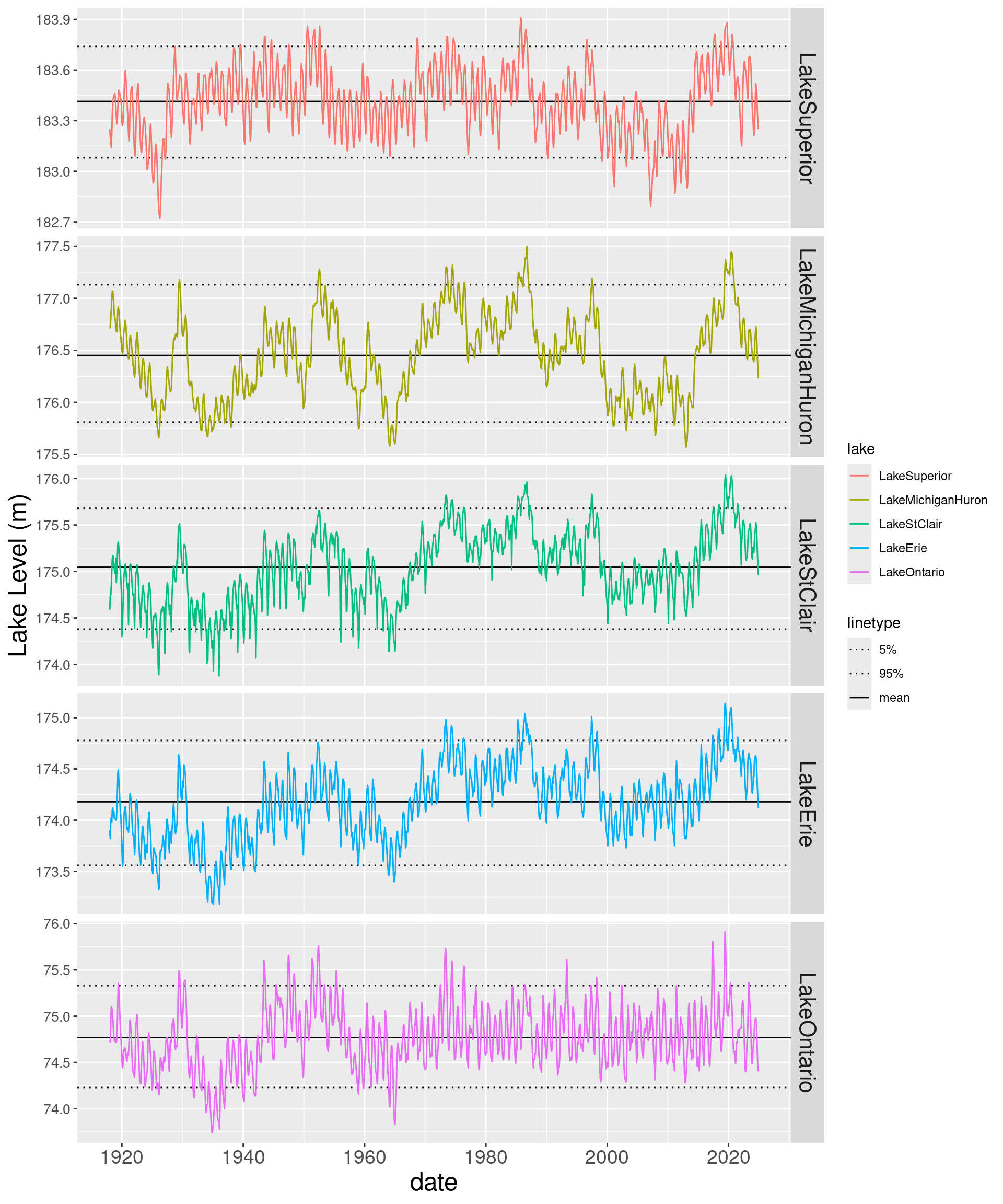
Wow, the lakes are back below their historic (2018 on) mean level. What are the driving factors behind the decrease from the recent peak period?
- less precipitation?
- more evaporation?
- less basin runoff?
- human controls?
- thermal contraction?
- some combination?
Citation
@online{isken2025,
author = {Isken, Mark},
title = {Plotting {Great} {Lakes} Water Level Data Using {R} - a 2025
Update},
date = {2025-02-09},
url = {https://bitsofanalytics.org//posts/great-lakes-water-levels-2024/get_plot_gl_water_levels.html},
langid = {en}
}